Slides
Two core issues remain: the spatial and the temporal. There are fewer well worked diagnostics for this than for the pure linear case owing to partial observability and/or coarse measurement in some fashion or other.
R
All of the computing will take place in the confines of pglm, a package in the spirit of plm for linear models. The help file provides an example of the familiar models. They are specified to match up with the syntax of R’s glm functions with family and link arguments.
# install.packages("pglm")
library(pglm)
## an ordered probit example
data('Fairness', package = 'pglm')
Parking <- subset(Fairness, good == 'parking')
op <- pglm(as.numeric(answer) ~ education + rule,
Parking[1:105, ],
family = ordinal('probit'), R = 5, print.level = 3,
method = 'bfgs', index = 'id', model = "random")
## a binomial (probit) example
data('UnionWage', package = 'pglm')
anb <- pglm(union ~ wage + exper + rural, UnionWage, family = binomial('probit'),
model = "pooling", method = "bfgs", print.level = 3, R = 5)
## a gaussian example on unbalanced panel data
data(Hedonic, package = "plm")
ra <- pglm(mv ~ crim + zn + indus + nox + age + rm, Hedonic, family = gaussian,
model = "random", print.level = 3, method = "nr", index = "townid")
## some count data models
data("PatentsRDUS", package="pglm")
la <- pglm(patents ~ lag(log(rd), 0:5) + scisect + log(capital72) + factor(year), PatentsRDUS,
family = negbin, model = "within", print.level = 3, method = "nr",
index = c('cusip', 'year'))
la <- pglm(patents ~ lag(log(rd), 0:5) + scisect + log(capital72) + factor(year), PatentsRDUS,
family = poisson, model = "pooling", index = c("cusip", "year"),
print.level = 0, method="nr")
## a tobit example
data("HealthIns", package="pglm")
HealthIns$med2 <- HealthIns$med / 1000
HealthIns2 <- HealthIns[-2209, ]
set.seed(2)
subs <- sample(1:20186, 200, replace = FALSE)
HealthIns2 <- HealthIns2[subs, ]
la <- pglm(med ~ mdu + disease + age, HealthIns2,
model = 'random', family = 'tobit', print.level = 0,
method = 'nr', R = 5)
Stata
Stata separates out the commands by family/link combination. So there are xtprobit and xtlogit for binary. xtoprobit for ordered probit models and xtologit [with random effects] for ordered outcomes.
Events
A Stata do file on the box is replicated here. First, a use of regular old glm.
library (haven)<- read_dta (url ("https://github.com/robertwwalker/Essex-Data/raw/main/Patents.dta" )).1 <- glm (PAT~ LOGR+ LOGR1+ LOGR2+ LOGR3+ LOGR4+ LOGR5+ dyear2+ dyear3+ dyear4+ dyear5, data= Patents, family= "poisson" )summary (Mod.1 )
Call:
glm(formula = PAT ~ LOGR + LOGR1 + LOGR2 + LOGR3 + LOGR4 + LOGR5 +
dyear2 + dyear3 + dyear4 + dyear5, family = "poisson", data = Patents)
Deviance Residuals:
Min 1Q Median 3Q Max
-24.1602 -3.0131 -1.6689 0.2972 23.3629
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.82831 0.01253 145.861 < 2e-16 ***
LOGR 0.15251 0.02939 5.189 2.11e-07 ***
LOGR1 0.02198 0.04083 0.538 0.590297
LOGR2 0.04369 0.03802 1.149 0.250557
LOGR3 0.08272 0.03568 2.318 0.020427 *
LOGR4 0.10397 0.03176 3.274 0.001061 **
LOGR5 0.30109 0.02063 14.594 < 2e-16 ***
dyear2 -0.04399 0.01320 -3.332 0.000862 ***
dyear3 -0.06043 0.01343 -4.499 6.81e-06 ***
dyear4 -0.18925 0.01368 -13.830 < 2e-16 ***
dyear5 -0.22979 0.01363 -16.854 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for poisson family taken to be 1)
Null deviance: 144500 on 1729 degrees of freedom
Residual deviance: 33745 on 1719 degrees of freedom
AIC: 39807
Number of Fisher Scoring iterations: 5
library (MASS).2 <- glm.nb (PAT~ LOGR+ LOGR1+ LOGR2+ LOGR3+ LOGR4+ LOGR5+ dyear2+ dyear3+ dyear4+ dyear5, data= Patents)summary (Mod.2 )
Call:
glm.nb(formula = PAT ~ LOGR + LOGR1 + LOGR2 + LOGR3 + LOGR4 +
LOGR5 + dyear2 + dyear3 + dyear4 + dyear5, data = Patents,
init.theta = 1.276776486, link = log)
Deviance Residuals:
Min 1Q Median 3Q Max
-2.6541 -1.0412 -0.3894 0.2827 4.3518
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.19343 0.05851 20.396 < 2e-16 ***
LOGR 0.40148 0.11591 3.464 0.000533 ***
LOGR1 -0.11198 0.15672 -0.714 0.474920
LOGR2 0.11039 0.14742 0.749 0.453957
LOGR3 0.08770 0.13696 0.640 0.521938
LOGR4 0.24389 0.12259 1.989 0.046657 *
LOGR5 0.17263 0.08478 2.036 0.041729 *
dyear2 -0.05346 0.07726 -0.692 0.488943
dyear3 -0.05482 0.07790 -0.704 0.481611
dyear4 -0.11764 0.07768 -1.514 0.129941
dyear5 -0.22006 0.07764 -2.835 0.004589 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for Negative Binomial(1.2768) family taken to be 1)
Null deviance: 7202.0 on 1729 degrees of freedom
Residual deviance: 1967.8 on 1719 degrees of freedom
AIC: 11594
Number of Fisher Scoring iterations: 1
Theta: 1.2768
Std. Err.: 0.0558
2 x log-likelihood: -11570.1870
Loading required package: maxLik
Loading required package: miscTools
Please cite the 'maxLik' package as:
Henningsen, Arne and Toomet, Ott (2011). maxLik: A package for maximum likelihood estimation in R. Computational Statistics 26(3), 443-458. DOI 10.1007/s00180-010-0217-1.
If you have questions, suggestions, or comments regarding the 'maxLik' package, please use a forum or 'tracker' at maxLik's R-Forge site:
https://r-forge.r-project.org/projects/maxlik/
Loading required package: plm
<- pdata.frame (Patents, index= c ("id" ,"YEAR" )).3 A <- pglm (PAT~ LOGR+ LOGR1+ LOGR2+ LOGR3+ LOGR4+ LOGR5+ dyear2+ dyear3+ dyear4+ dyear5+ LOGK+ SCISECT, Patents.pdata, effect= "individual" , model= "random" , family= "poisson" )summary (Mod.3 A)
--------------------------------------------
Maximum Likelihood estimation
Newton-Raphson maximisation, 4 iterations
Return code 8: successive function values within relative tolerance limit (reltol)
Log-Likelihood: -5234.927
14 free parameters
Estimates:
Estimate Std. error t value Pr(> t)
(Intercept) 0.41079 0.14674 2.799 0.005121 **
LOGR 0.40345 0.04350 9.274 < 2e-16 ***
LOGR1 -0.04618 0.04822 -0.958 0.338277
LOGR2 0.10792 0.04471 2.414 0.015788 *
LOGR3 0.02977 0.04132 0.720 0.471221
LOGR4 0.01070 0.03771 0.284 0.776679
LOGR5 0.04061 0.03157 1.286 0.198364
dyear2 -0.04496 0.01313 -3.425 0.000616 ***
dyear3 -0.04839 0.01340 -3.610 0.000306 ***
dyear4 -0.17416 0.01397 -12.467 < 2e-16 ***
dyear5 -0.22590 0.01466 -15.404 < 2e-16 ***
LOGK 0.29169 0.03934 7.415 1.21e-13 ***
SCISECT 0.25700 0.11227 2.289 0.022074 *
sigma 1.16969 0.09471 12.350 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
--------------------------------------------
.3 B <- pglm (PAT~ LOGR+ LOGR1+ LOGR2+ LOGR3+ LOGR4+ LOGR5+ dyear2+ dyear3+ dyear4+ dyear5, Patents.pdata, effect= "individual" , model= "random" , family= "poisson" )summary (Mod.3 B)
--------------------------------------------
Maximum Likelihood estimation
Newton-Raphson maximisation, 5 iterations
Return code 1: gradient close to zero (gradtol)
Log-Likelihood: -5263.611
12 free parameters
Estimates:
Estimate Std. error t value Pr(> t)
(Intercept) 1.402861 0.067052 20.922 < 2e-16 ***
LOGR 0.476575 0.042263 11.276 < 2e-16 ***
LOGR1 -0.007708 0.047934 -0.161 0.872242
LOGR2 0.136413 0.044734 3.049 0.002293 **
LOGR3 0.059191 0.041279 1.434 0.151598
LOGR4 0.027518 0.037601 0.732 0.464267
LOGR5 0.082547 0.030977 2.665 0.007704 **
dyear2 -0.046879 0.013131 -3.570 0.000357 ***
dyear3 -0.056088 0.013362 -4.198 2.7e-05 ***
dyear4 -0.190312 0.013789 -13.802 < 2e-16 ***
dyear5 -0.252677 0.014197 -17.798 < 2e-16 ***
sigma 1.154854 0.094192 12.261 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
--------------------------------------------
.3 C <- pglm (PAT~ LOGR+ LOGR1+ LOGR2+ LOGR3+ LOGR4+ LOGR5+ dyear2+ dyear3+ dyear4+ dyear5+ LOGK+ SCISECT-1 , Patents.pdata, effect= "individual" , model= "random" , family= "poisson" )summary (Mod.3 C)
--------------------------------------------
Maximum Likelihood estimation
Newton-Raphson maximisation, 4 iterations
Return code 1: gradient close to zero (gradtol)
Log-Likelihood: -5238.845
13 free parameters
Estimates:
Estimate Std. error t value Pr(> t)
LOGR 0.382042 0.042877 8.910 < 2e-16 ***
LOGR1 -0.055620 0.048157 -1.155 0.248098
LOGR2 0.100308 0.044621 2.248 0.024576 *
LOGR3 0.020949 0.041203 0.508 0.611146
LOGR4 0.005334 0.037709 0.141 0.887523
LOGR5 0.028364 0.031336 0.905 0.365383
dyear2 -0.042732 0.013109 -3.260 0.001115 **
dyear3 -0.044442 0.013332 -3.333 0.000858 ***
dyear4 -0.167871 0.013796 -12.168 < 2e-16 ***
dyear5 -0.216749 0.014303 -15.154 < 2e-16 ***
LOGK 0.388567 0.019946 19.481 < 2e-16 ***
SCISECT 0.419111 0.097521 4.298 1.73e-05 ***
sigma 1.117773 0.088520 12.627 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
--------------------------------------------
.4 A <- pglm (PAT~ LOGR+ LOGR1+ LOGR2+ LOGR3+ LOGR4+ LOGR5+ dyear2+ dyear3+ dyear4+ dyear5+ LOGK+ SCISECT, Patents.pdata, effect= "individual" , model= "random" , family= "negbin" )summary (Mod.4 A)
--------------------------------------------
Maximum Likelihood estimation
Newton-Raphson maximisation, 6 iterations
Return code 1: gradient close to zero (gradtol)
Log-Likelihood: -4948.494
15 free parameters
Estimates:
Estimate Std. error t value Pr(> t)
(Intercept) 0.899562 0.168111 5.351 8.75e-08 ***
LOGR 0.350312 0.065282 5.366 8.04e-08 ***
LOGR1 -0.003032 0.075092 -0.040 0.967796
LOGR2 0.104988 0.068849 1.525 0.127284
LOGR3 0.016352 0.063638 0.257 0.797210
LOGR4 0.035942 0.058716 0.612 0.540445
LOGR5 0.071832 0.048289 1.488 0.136867
dyear2 -0.043674 0.021343 -2.046 0.040734 *
dyear3 -0.055660 0.021857 -2.547 0.010881 *
dyear4 -0.183105 0.022718 -8.060 7.64e-16 ***
dyear5 -0.230044 0.023152 -9.936 < 2e-16 ***
LOGK 0.161937 0.041787 3.875 0.000107 ***
SCISECT 0.117642 0.106616 1.103 0.269848
a 2.685210 0.258163 10.401 < 2e-16 ***
b 2.015688 0.217631 9.262 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
--------------------------------------------
.4 B <- pglm (PAT~ LOGR+ LOGR1+ LOGR2+ LOGR3+ LOGR4+ LOGR5+ dyear2+ dyear3+ dyear4+ dyear5, Patents.pdata, effect= "individual" , model= "random" , family= "negbin" )summary (Mod.4 B)
--------------------------------------------
Maximum Likelihood estimation
Newton-Raphson maximisation, 6 iterations
Return code 1: gradient close to zero (gradtol)
Log-Likelihood: -4956.259
13 free parameters
Estimates:
Estimate Std. error t value Pr(> t)
(Intercept) 1.39944 0.10060 13.910 < 2e-16 ***
LOGR 0.38762 0.06492 5.970 2.37e-09 ***
LOGR1 0.01833 0.07597 0.241 0.80935
LOGR2 0.12909 0.06972 1.852 0.06408 .
LOGR3 0.02469 0.06446 0.383 0.70172
LOGR4 0.05775 0.05873 0.983 0.32544
LOGR5 0.10003 0.04750 2.106 0.03521 *
dyear2 -0.04671 0.02180 -2.142 0.03219 *
dyear3 -0.06512 0.02223 -2.930 0.00339 **
dyear4 -0.20006 0.02282 -8.765 < 2e-16 ***
dyear5 -0.25096 0.02298 -10.921 < 2e-16 ***
a 2.64396 0.25190 10.496 < 2e-16 ***
b 2.02183 0.21889 9.237 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
--------------------------------------------
.4 C <- pglm (PAT~ LOGR+ LOGR1+ LOGR2+ LOGR3+ LOGR4+ LOGR5+ dyear2+ dyear3+ dyear4+ dyear5+ LOGK+ SCISECT-1 , Patents.pdata, effect= "individual" , model= "random" , family= "negbin" )summary (Mod.4 C)
--------------------------------------------
Maximum Likelihood estimation
Newton-Raphson maximisation, 6 iterations
Return code 1: gradient close to zero (gradtol)
Log-Likelihood: -4963.305
14 free parameters
Estimates:
Estimate Std. error t value Pr(> t)
LOGR 0.296803 0.066943 4.434 9.26e-06 ***
LOGR1 -0.005170 0.078300 -0.066 0.9474
LOGR2 0.085684 0.071482 1.199 0.2307
LOGR3 0.004177 0.065959 0.063 0.9495
LOGR4 0.010518 0.060898 0.173 0.8629
LOGR5 0.048459 0.050342 0.963 0.3357
dyear2 -0.032498 0.021569 -1.507 0.1319
dyear3 -0.037854 0.021894 -1.729 0.0838 .
dyear4 -0.159944 0.022586 -7.082 1.42e-12 ***
dyear5 -0.201171 0.022791 -8.827 < 2e-16 ***
LOGK 0.343399 0.027143 12.652 < 2e-16 ***
SCISECT 0.427951 0.089606 4.776 1.79e-06 ***
a 2.435387 0.225784 10.786 < 2e-16 ***
b 2.154018 0.240574 8.954 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
--------------------------------------------
The GEE
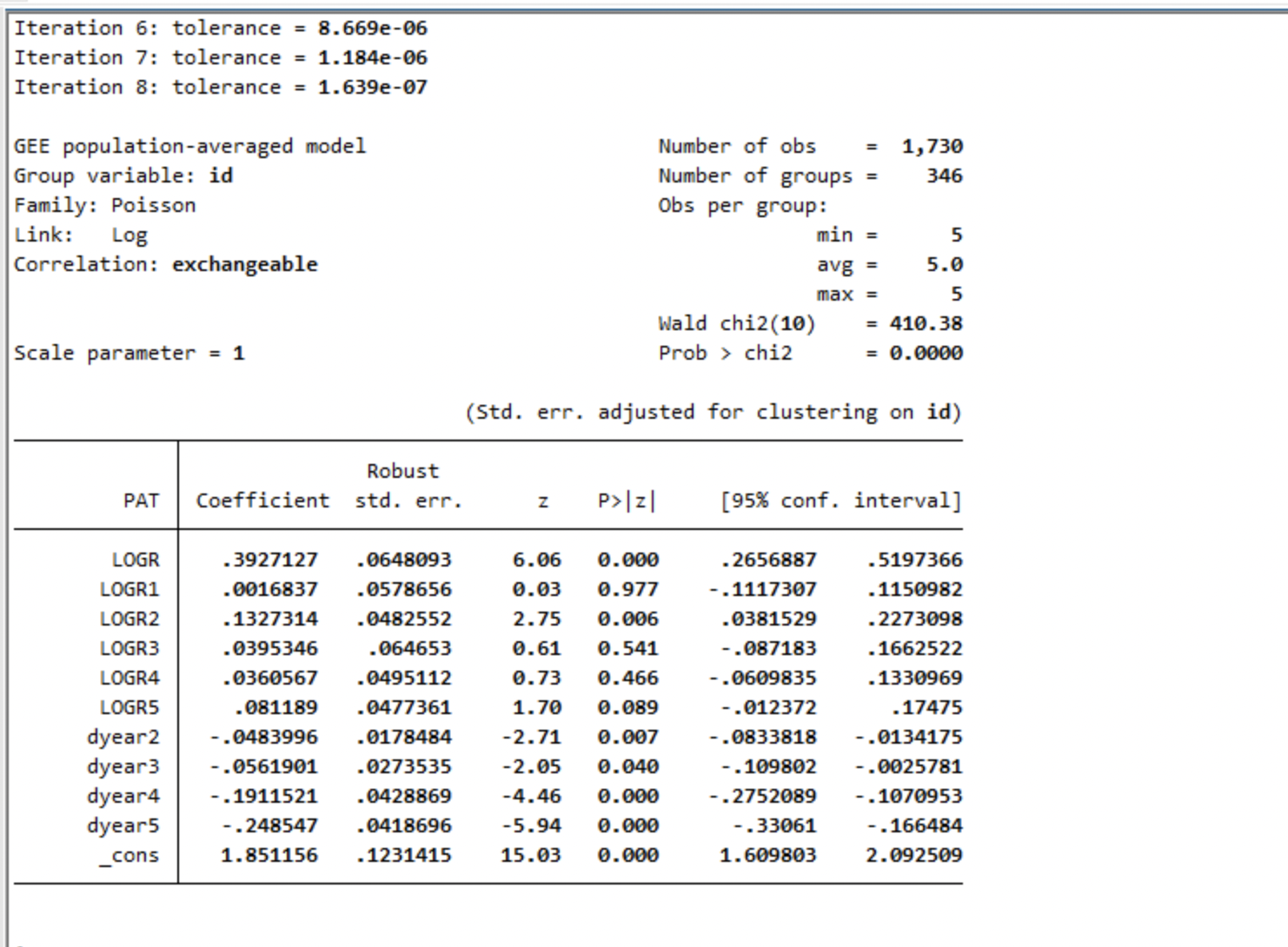
library (geepack)<- geeglm (PAT~ LOGR+ LOGR1+ LOGR2+ LOGR3+ LOGR4+ LOGR5+ dyear2+ dyear3+ dyear4+ dyear5, data= Patents, id= id, corstr= "exchangeable" , family= poisson)summary (gee.Mod)
Call:
geeglm(formula = PAT ~ LOGR + LOGR1 + LOGR2 + LOGR3 + LOGR4 +
LOGR5 + dyear2 + dyear3 + dyear4 + dyear5, family = poisson,
data = Patents, id = id, corstr = "exchangeable")
Coefficients:
Estimate Std.err Wald Pr(>|W|)
(Intercept) 1.851156 0.122963 226.638 < 2e-16 ***
LOGR 0.392713 0.064716 36.824 1.29e-09 ***
LOGR1 0.001683 0.057782 0.001 0.97677
LOGR2 0.132732 0.048185 7.588 0.00588 **
LOGR3 0.039535 0.064560 0.375 0.54029
LOGR4 0.036056 0.049440 0.532 0.46582
LOGR5 0.081190 0.047667 2.901 0.08851 .
dyear2 -0.048400 0.017823 7.375 0.00661 **
dyear3 -0.056190 0.027314 4.232 0.03967 *
dyear4 -0.191152 0.042825 19.923 8.06e-06 ***
dyear5 -0.248547 0.041809 35.341 2.77e-09 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation structure = exchangeable
Estimated Scale Parameters:
Estimate Std.err
(Intercept) 22.57 4.102
Link = identity
Estimated Correlation Parameters:
Estimate Std.err
alpha 0.8983 0.06245
Number of clusters: 346 Maximum cluster size: 5
<- geeglm (PAT~ LOGR+ LOGR1+ LOGR2+ LOGR3+ LOGR4+ LOGR5+ dyear2+ dyear3+ dyear4+ dyear5, data= Patents, id= id, corstr= "ar1" , family= poisson)summary (gee.Mod)
Call:
geeglm(formula = PAT ~ LOGR + LOGR1 + LOGR2 + LOGR3 + LOGR4 +
LOGR5 + dyear2 + dyear3 + dyear4 + dyear5, family = poisson,
data = Patents, id = id, corstr = "ar1")
Coefficients:
Estimate Std.err Wald Pr(>|W|)
(Intercept) 1.8121 0.1270 203.72 < 2e-16 ***
LOGR 0.3096 0.0554 31.17 2.4e-08 ***
LOGR1 0.0254 0.0549 0.21 0.6435
LOGR2 0.1624 0.0518 9.82 0.0017 **
LOGR3 0.0801 0.0564 2.02 0.1555
LOGR4 0.0793 0.0446 3.16 0.0756 .
LOGR5 0.0408 0.0411 0.99 0.3200
dyear2 -0.0494 0.0175 8.01 0.0047 **
dyear3 -0.0542 0.0266 4.14 0.0419 *
dyear4 -0.1817 0.0427 18.08 2.1e-05 ***
dyear5 -0.2333 0.0409 32.48 1.2e-08 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation structure = ar1
Estimated Scale Parameters:
Estimate Std.err
(Intercept) 22.3 4.02
Link = identity
Estimated Correlation Parameters:
Estimate Std.err
alpha 0.948 0.0265
Number of clusters: 346 Maximum cluster size: 5
And an anova…
Analysis of 'Wald statistic' Table
Model: poisson, link: log
Response: PAT
Terms added sequentially (first to last)
Df X2 P(>|Chi|)
LOGR 1 368 < 2e-16 ***
LOGR1 1 23 1.9e-06 ***
LOGR2 1 19 1.4e-05 ***
LOGR3 1 16 5.8e-05 ***
LOGR4 1 7 0.00708 **
LOGR5 1 1 0.28590
dyear2 1 4 0.05740 .
dyear3 1 11 0.00099 ***
dyear4 1 0 0.67359
dyear5 1 32 1.2e-08 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
BKT and Carter-Signorino
Load the data
<- read_dta (url ("https://github.com/robertwwalker/Essex-Data/raw/main/bkt98ajps.dta" )).1 <- glm (dispute~ dem+ growth+ allies+ contig+ capratio+ trade, family= binomial ("logit" ), data= BKT.data)summary (Mod.1 )
Call:
glm(formula = dispute ~ dem + growth + allies + contig + capratio +
trade, family = binomial("logit"), data = BKT.data)
Deviance Residuals:
Min 1Q Median 3Q Max
-0.748 -0.319 -0.261 -0.152 4.096
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -3.29e+00 7.92e-02 -41.54 < 2e-16 ***
dem -4.97e-02 7.38e-03 -6.73 1.7e-11 ***
growth -2.23e-02 8.52e-03 -2.62 0.0088 **
allies -8.21e-01 8.00e-02 -10.26 < 2e-16 ***
contig 1.31e+00 7.96e-02 16.49 < 2e-16 ***
capratio -3.07e-03 4.17e-04 -7.37 1.8e-13 ***
trade -6.61e+01 1.34e+01 -4.92 8.6e-07 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 7719.2 on 20989 degrees of freedom
Residual deviance: 6955.1 on 20983 degrees of freedom
AIC: 6969
Number of Fisher Scoring iterations: 8
.1 B <- glm (dispute~ dem+ growth+ allies+ contig+ capratio+ trade+ as.factor (py), family= binomial ("logit" ), data= BKT.data)
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Call:
glm(formula = dispute ~ dem + growth + allies + contig + capratio +
trade + as.factor(py), family = binomial("logit"), data = BKT.data)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.362 -0.225 -0.140 -0.080 3.988
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -9.43e-01 9.28e-02 -10.16 < 2e-16 ***
dem -5.47e-02 8.00e-03 -6.84 8.0e-12 ***
growth -1.15e-02 9.24e-03 -1.24 0.21492
allies -4.71e-01 9.00e-02 -5.23 1.7e-07 ***
contig 6.99e-01 8.93e-02 7.82 5.3e-15 ***
capratio -3.03e-03 4.17e-04 -7.28 3.3e-13 ***
trade -1.27e+01 1.05e+01 -1.21 0.22735
as.factor(py)2 -2.04e+00 1.41e-01 -14.48 < 2e-16 ***
as.factor(py)3 -2.15e+00 1.54e-01 -13.92 < 2e-16 ***
as.factor(py)4 -2.37e+00 1.74e-01 -13.64 < 2e-16 ***
as.factor(py)5 -2.62e+00 2.01e-01 -13.04 < 2e-16 ***
as.factor(py)6 -3.09e+00 2.52e-01 -12.25 < 2e-16 ***
as.factor(py)7 -3.08e+00 2.52e-01 -12.21 < 2e-16 ***
as.factor(py)8 -3.00e+00 2.46e-01 -12.23 < 2e-16 ***
as.factor(py)9 -2.95e+00 2.46e-01 -11.98 < 2e-16 ***
as.factor(py)10 -3.24e+00 2.86e-01 -11.32 < 2e-16 ***
as.factor(py)11 -3.45e+00 3.24e-01 -10.66 < 2e-16 ***
as.factor(py)12 -3.32e+00 3.10e-01 -10.72 < 2e-16 ***
as.factor(py)13 -3.92e+00 4.14e-01 -9.46 < 2e-16 ***
as.factor(py)14 -4.56e+00 5.82e-01 -7.84 4.6e-15 ***
as.factor(py)15 -4.58e+00 5.82e-01 -7.87 3.5e-15 ***
as.factor(py)16 -3.45e+00 3.41e-01 -10.12 < 2e-16 ***
as.factor(py)17 -4.22e+00 5.05e-01 -8.35 < 2e-16 ***
as.factor(py)18 -5.59e+00 1.00e+00 -5.57 2.5e-08 ***
as.factor(py)19 -3.61e+00 3.85e-01 -9.36 < 2e-16 ***
as.factor(py)20 -3.95e+00 4.53e-01 -8.71 < 2e-16 ***
as.factor(py)21 -4.80e+00 7.11e-01 -6.74 1.5e-11 ***
as.factor(py)22 -4.31e+00 5.82e-01 -7.39 1.4e-13 ***
as.factor(py)23 -4.13e+00 5.83e-01 -7.08 1.4e-12 ***
as.factor(py)24 -3.99e+00 5.83e-01 -6.84 7.7e-12 ***
as.factor(py)25 -1.76e+01 3.31e+02 -0.05 0.95752
as.factor(py)26 -2.97e+00 4.19e-01 -7.10 1.2e-12 ***
as.factor(py)27 -1.76e+01 3.85e+02 -0.05 0.96353
as.factor(py)28 -1.76e+01 3.83e+02 -0.05 0.96347
as.factor(py)29 -3.92e+00 7.14e-01 -5.49 3.9e-08 ***
as.factor(py)30 -3.42e+00 5.86e-01 -5.84 5.2e-09 ***
as.factor(py)31 -4.41e+00 1.01e+00 -4.39 1.1e-05 ***
as.factor(py)32 -3.57e+00 7.15e-01 -4.99 6.0e-07 ***
as.factor(py)33 -4.07e+00 1.01e+00 -4.05 5.2e-05 ***
as.factor(py)34 -3.31e+00 7.16e-01 -4.61 3.9e-06 ***
as.factor(py)35 -3.83e+00 1.01e+00 -3.81 0.00014 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 7719.2 on 20989 degrees of freedom
Residual deviance: 5109.4 on 20949 degrees of freedom
AIC: 5191
Number of Fisher Scoring iterations: 17
.1 C <- glm (dispute~ dem+ growth+ allies+ contig+ capratio+ trade+ py+ pys1+ pys2+ pys3, family= binomial ("logit" ), data= BKT.data)summary (Mod.1 C)
Call:
glm(formula = dispute ~ dem + growth + allies + contig + capratio +
trade + py + pys1 + pys2 + pys3, family = binomial("logit"),
data = BKT.data)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.349 -0.221 -0.138 -0.092 4.063
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 8.50e-01 1.60e-01 5.31 1.1e-07 ***
dem -5.46e-02 7.97e-03 -6.84 7.7e-12 ***
growth -1.15e-02 9.20e-03 -1.25 0.21
allies -4.70e-01 8.96e-02 -5.25 1.5e-07 ***
contig 6.94e-01 8.90e-02 7.79 6.6e-15 ***
capratio -3.04e-03 4.17e-04 -7.29 3.0e-13 ***
trade -1.29e+01 1.05e+01 -1.23 0.22
py -1.82e+00 1.11e-01 -16.30 < 2e-16 ***
pys1 -2.45e-01 2.61e-02 -9.37 < 2e-16 ***
pys2 7.92e-02 1.09e-02 7.24 4.5e-13 ***
pys3 -1.09e-02 2.76e-03 -3.96 7.5e-05 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 7719.2 on 20989 degrees of freedom
Residual deviance: 5165.8 on 20979 degrees of freedom
AIC: 5188
Number of Fisher Scoring iterations: 8
.2 A <- glm (dispute~ dem+ growth+ allies+ contig+ capratio+ trade, family= binomial ("logit" ), subset = contdisp!= 1 , data= BKT.data)summary (Mod.2 A)
Call:
glm(formula = dispute ~ dem + growth + allies + contig + capratio +
trade, family = binomial("logit"), data = BKT.data, subset = contdisp !=
1)
Deviance Residuals:
Min 1Q Median 3Q Max
-0.520 -0.218 -0.169 -0.120 3.868
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -4.33e+00 1.15e-01 -37.78 < 2e-16 ***
dem -4.01e-02 1.01e-02 -3.99 6.7e-05 ***
growth -3.43e-02 1.25e-02 -2.74 0.0062 **
allies -4.80e-01 1.13e-01 -4.25 2.1e-05 ***
contig 1.35e+00 1.21e-01 11.20 < 2e-16 ***
capratio -1.96e-03 5.01e-04 -3.92 9.0e-05 ***
trade -2.11e+01 1.13e+01 -1.86 0.0623 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 3978.5 on 20447 degrees of freedom
Residual deviance: 3693.8 on 20441 degrees of freedom
AIC: 3708
Number of Fisher Scoring iterations: 9
.2 B <- glm (dispute~ dem+ growth+ allies+ contig+ capratio+ trade+ py+ pys1+ pys2+ pys3, family= binomial ("logit" ), subset = contdisp!= 1 , data= BKT.data)summary (Mod.2 B)
Call:
glm(formula = dispute ~ dem + growth + allies + contig + capratio +
trade + py + pys1 + pys2 + pys3, family = binomial("logit"),
data = BKT.data, subset = contdisp != 1)
Deviance Residuals:
Min 1Q Median 3Q Max
-0.734 -0.210 -0.135 -0.098 3.732
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -3.956674 0.297828 -13.29 < 2e-16 ***
dem -0.038809 0.010046 -3.86 0.00011 ***
growth -0.040144 0.012502 -3.21 0.00132 **
allies -0.365957 0.114272 -3.20 0.00136 **
contig 0.993251 0.124133 8.00 1.2e-15 ***
capratio -0.002289 0.000523 -4.38 1.2e-05 ***
trade -3.809220 9.679795 -0.39 0.69393
py 0.390965 0.157920 2.48 0.01330 *
pys1 0.089719 0.031787 2.82 0.00477 **
pys2 -0.028888 0.012634 -2.29 0.02222 *
pys3 0.003314 0.002974 1.11 0.26513
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 3978.5 on 20447 degrees of freedom
Residual deviance: 3502.8 on 20437 degrees of freedom
AIC: 3525
Number of Fisher Scoring iterations: 9
.3 A <- glm (dispute~ dem+ growth+ allies+ contig+ capratio+ trade+ py+ pys1+ pys2+ pys3, family= binomial ("logit" ), subset = prefail< 1 , data= BKT.data)summary (Mod.3 A)
Call:
glm(formula = dispute ~ dem + growth + allies + contig + capratio +
trade + py + pys1 + pys2 + pys3, family = binomial("logit"),
data = BKT.data, subset = prefail < 1)
Deviance Residuals:
Min 1Q Median 3Q Max
-0.682 -0.159 -0.109 -0.081 3.856
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -2.132791 0.369090 -5.78 7.5e-09 ***
dem -0.046143 0.013330 -3.46 0.00054 ***
growth -0.022954 0.017754 -1.29 0.19604
allies -0.423113 0.163691 -2.58 0.00974 **
contig 1.105361 0.172537 6.41 1.5e-10 ***
capratio -0.001916 0.000601 -3.19 0.00142 **
trade -3.553603 11.726412 -0.30 0.76186
py -1.081689 0.235688 -4.59 4.4e-06 ***
pys1 -0.183730 0.050096 -3.67 0.00024 ***
pys2 0.069295 0.019788 3.50 0.00046 ***
pys3 -0.013987 0.004484 -3.12 0.00181 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 2183.3 on 16990 degrees of freedom
Residual deviance: 1928.1 on 16980 degrees of freedom
AIC: 1950
Number of Fisher Scoring iterations: 9
Carter and Signorino
Use a Taylor series to clean this up with a suggestion that no baseline hazard is more than cubic [two inflection points].
<- glm (dispute~ dem+ growth+ allies+ contig+ capratio+ trade+ prefail+ py+ I (py^ 2 )+ I (py^ 3 ), family= binomial ("logit" ), data= BKT.data)summary (Mod.CS)
Call:
glm(formula = dispute ~ dem + growth + allies + contig + capratio +
trade + prefail + py + I(py^2) + I(py^3), family = binomial("logit"),
data = BKT.data)
Deviance Residuals:
Min 1Q Median 3Q Max
-2.576 -0.192 -0.128 -0.092 3.754
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -1.14e+00 1.18e-01 -9.64 < 2e-16 ***
dem -4.09e-02 8.23e-03 -4.97 6.7e-07 ***
growth -2.44e-02 9.55e-03 -2.56 0.0105 *
allies -2.67e-01 9.20e-02 -2.90 0.0038 **
contig 6.62e-01 9.20e-02 7.19 6.4e-13 ***
capratio -2.00e-03 3.71e-04 -5.40 6.6e-08 ***
trade -1.06e+01 1.04e+01 -1.03 0.3049
prefail 1.75e-01 8.96e-03 19.53 < 2e-16 ***
py -7.48e-01 4.14e-02 -18.08 < 2e-16 ***
I(py^2) 4.22e-02 3.72e-03 11.34 < 2e-16 ***
I(py^3) -7.18e-04 8.76e-05 -8.19 2.5e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 7719.2 on 20989 degrees of freedom
Residual deviance: 4909.5 on 20979 degrees of freedom
AIC: 4932
Number of Fisher Scoring iterations: 8
As a technical matter, none of this is exactly equivalent to a Cox model. That requires the cloglog.
<- glm (dispute~ dem+ growth+ allies+ contig+ capratio+ trade+ prefail+ py+ I (py^ 2 )+ I (py^ 3 ), family= binomial ("cloglog" ), data= BKT.data)summary (Mod.CS)
Call:
glm(formula = dispute ~ dem + growth + allies + contig + capratio +
trade + prefail + py + I(py^2) + I(py^3), family = binomial("cloglog"),
data = BKT.data)
Deviance Residuals:
Min 1Q Median 3Q Max
-3.396 -0.194 -0.134 -0.097 3.700
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -1.13e+00 1.07e-01 -10.62 < 2e-16 ***
dem -4.16e-02 7.50e-03 -5.54 2.9e-08 ***
growth -1.97e-02 8.00e-03 -2.47 0.0137 *
allies -2.33e-01 7.99e-02 -2.92 0.0035 **
contig 5.16e-01 7.86e-02 6.56 5.4e-11 ***
capratio -2.13e-03 3.60e-04 -5.93 3.0e-09 ***
trade -1.15e+01 1.01e+01 -1.13 0.2567
prefail 1.15e-01 6.46e-03 17.80 < 2e-16 ***
py -7.03e-01 3.84e-02 -18.32 < 2e-16 ***
I(py^2) 3.89e-02 3.51e-03 11.07 < 2e-16 ***
I(py^3) -6.55e-04 8.33e-05 -7.85 4.1e-15 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 7719.2 on 20989 degrees of freedom
Residual deviance: 4944.0 on 20979 degrees of freedom
AIC: 4966
Number of Fisher Scoring iterations: 9